Paper Information
Citation: Zhao, Z., Chen, B., Li, J., Chen, L., Wen, L., Wang, P., Zhu, Z., Zhang, D., Wan, Z., Li, Y., Dai, Z., Chen, X., & Yu, K. (2024). ChemDFM-X: Towards Large Multimodal Model for Chemistry. Science China Information Sciences, 67(12), 220109. https://doi.org/10.1007/s11432-024-4243-0
Publication: Science China Information Sciences, December 2024
Additional Resources:
- arXiv Version
- Code Repository (if available)
What kind of paper is this?
This is primarily a Method paper with a significant Resource contribution.
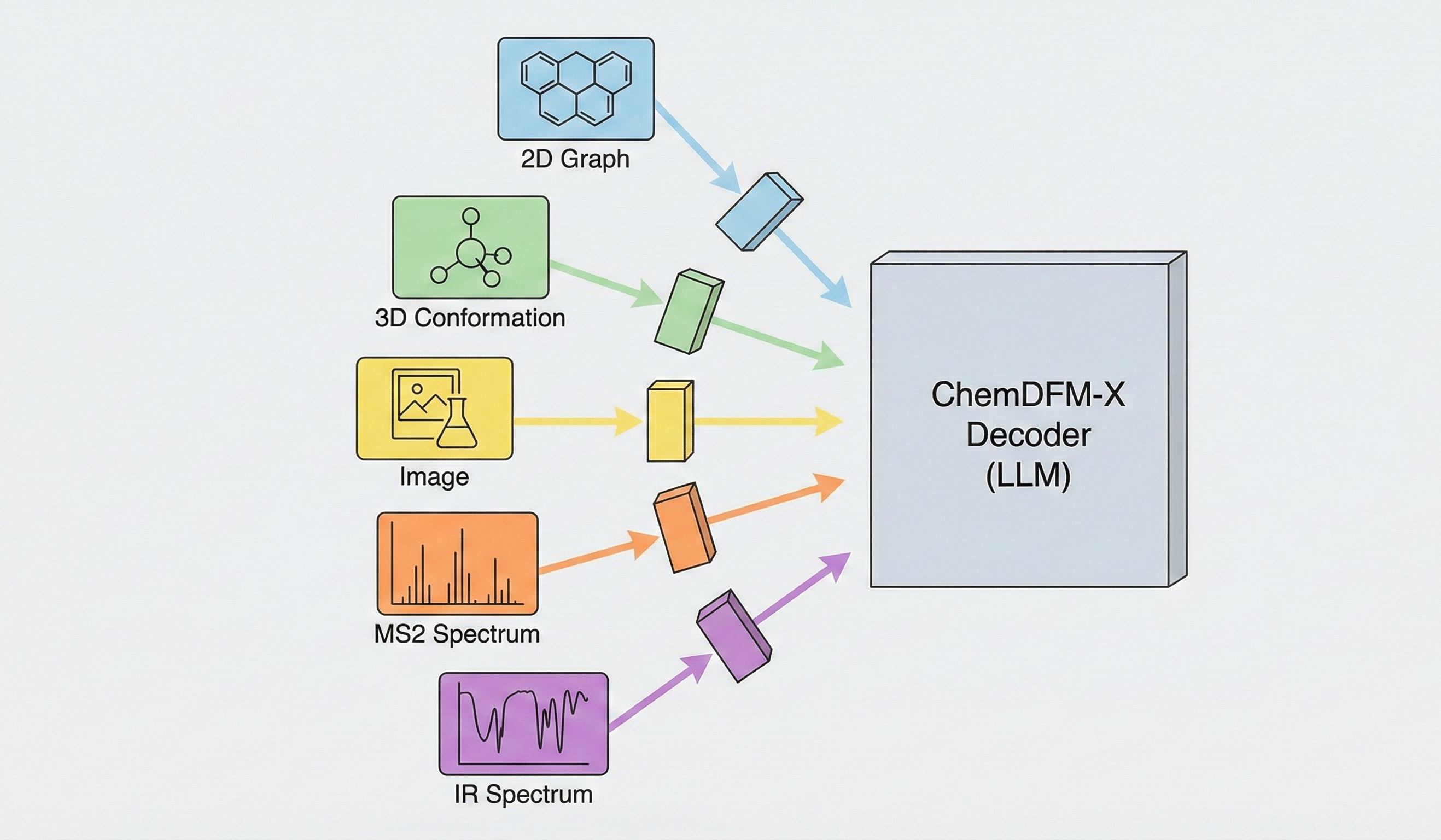
Method: The paper proposes a novel “Cross-modal Dialogue Foundation Model” architecture that aligns five distinct chemical modalities (2D graphs, 3D conformations, images, MS2 spectra, IR spectra) to a single LLM decoder using separate encoders and projection modules. It demonstrates state-of-the-art generalist performance through extensive baseline comparisons.
Resource: The paper addresses the scarcity of multimodal chemical data by constructing a 7.6M instruction-tuning dataset. This dataset is largely synthesized from seed SMILES strings using approximate calculations (MMFF94, CFM-ID, Chemprop-IR) and specialist model predictions.
What is the motivation?
Current AI for chemistry suffers from a fundamental dichotomy:
- Specialist Models: Achieve high performance on single tasks (e.g., property prediction, molecular generation) but lack generalization capabilities and natural language interaction.
- Generalist LMMs: Existing Chemical LLMs (like ChemDFM v1 or ChemLLM) are limited to text and SMILES modalities. General domain LMMs (like GPT-4V) lack deep chemical domain knowledge.
The authors identify a specific need for Chemical General Intelligence (CGI) that can interpret the diverse modalities chemists actually use in practice - particularly experimental characterization data like mass spectrometry (MS2) and infrared (IR) spectra, which are often ignored in favor of pure structural representations.
What is the novelty here?
The core novelty lies in the “Any-to-Text” alignment strategy via synthetic data scaling:
Comprehensive Modality Support: One of the first models to support MS2 and IR spectra alongside 2D graphs, 3D conformations, and images in a dialogue format. This enables interpretation of both experimental characterization data and molecular structures.
Synthetic Data Generation Pipeline: The authors generated a 7.6M sample dataset by starting with 1.3M seed SMILES and using “approximate calculations” to generate missing modalities:
- 3D conformations via MMFF94 force field optimization
- MS2 spectra via CFM-ID 4.0 (Competitive Fragmentation Modeling)
- IR spectra via Chemprop-IR (Message Passing Neural Network)
Cross-Modal Synergy: The model demonstrates that training on reaction images improves recognition performance by leveraging semantic chemical knowledge (reaction rules) to correct visual recognition errors - an emergent capability from multimodal training.
What experiments were performed?
The model was evaluated using a customized version of ChemLLMBench and MoleculeNet across three modality categories:
Structural Modalities (2D Graphs & 3D Conformations):
- Molecule recognition and captioning
- Property prediction (MoleculeNet: BACE, BBBP, HIV, Tox21, etc.)
- Compared against specialist models (Mole-BERT, Uni-Mol) and generalist models (3D-MOLM)
Visual Modalities (Images):
- Single molecule image recognition
- Reaction image recognition
- Compared against GPT-4V, Gemini 1.5 Pro, Qwen-VL, and MolScribe
Characterization Modalities (MS2 & IR Spectra):
- Spectral analysis tasks (identifying molecules from spectra)
- Contextualized spectral interpretation (combining spectra with reaction context)
- Novel evaluation requiring integration of spectroscopic data with reaction knowledge
What were the outcomes and conclusions drawn?
Key Findings:
SOTA Generalist Performance: ChemDFM-X outperformed existing generalist models (like 3D-MOLM, ChemLLM) and achieved performance comparable to specialist models on several tasks, validating the multimodal training approach.
Failure of General LMMs: General vision models (GPT-4V, Gemini 1.5 Pro) failed significantly on chemical image recognition tasks (0% accuracy on some baselines), demonstrating that chemical domain knowledge cannot be assumed from general pre-training.
Cross-Modal Error Correction: In reaction image recognition, ChemDFM-X achieved higher accuracy (65.1%) than on single molecules (57.6%). The authors conclude the model uses its internal knowledge of chemical reaction rules to “auto-correct” recognition errors in the visual modality - an emergent capability from multimodal training.
Spectral Understanding: The model successfully combined spectral data (MS2/IR) with reaction context (known reactants/products) to identify unknown molecules, demonstrating practical utility for experimental chemistry workflows. This capability was previously restricted to highly specialized analytical tools.
Main Conclusion: The “separate encoders + unified decoder” architecture with synthetic data generation enables effective multimodal chemical understanding, bridging the gap between specialist and generalist AI systems for chemistry.
Reproducibility Details
Data
The authors constructed a 7.6M sample instruction-tuning dataset derived from 1.3M seed SMILES (sourced from PubChem and USPTO).
Generation Pipeline:
| Modality | Generation Method | Tool/Model | Sample Count |
|---|---|---|---|
| 2D Graphs | Direct extraction from SMILES | RDKit | 1.1M |
| 3D Conformations | Force field optimization | RDKit + MMFF94 | 1.3M (pseudo-optimal) |
| Molecule Images | Rendering with augmentation | RDKit, Indigo, ChemPix | ~1M (including handwritten style) |
| Reaction Images | Rendering from reaction SMILES | RDKit | 300K |
| MS2 Spectra | Computational prediction | CFM-ID 4.0 | ~700K |
| IR Spectra | Computational prediction | Chemprop-IR | ~1M |
Data Augmentation:
- Molecule images augmented with “handwritten” style using the ChemPix pipeline
- Multiple rendering styles (RDKit default, Indigo clean)
- Spectra generated at multiple energy levels (10eV, 20eV, 40eV for MS2)
Algorithms
Architecture: “Separate Encoders + Unified Decoder”
Modality Alignment:
- Each modality has a dedicated encoder (frozen pre-trained models where available)
- Simple 2-layer MLP projector (Linear → GELU → Linear) maps encoder features to LLM input space
- All projected features are concatenated and fed to unified LLM decoder
Image Handling:
- H-Reducer module compresses image tokens by factor of $n=8$ to handle high-resolution chemical images that exceed standard CLIP token limits
- Enables processing of complex reaction schemes and detailed molecular depictions
Models
Base LLM:
- ChemDFM (13B): LLaMA-based model pre-trained on chemical text and SMILES
Modality Encoders:
| Modality | Encoder | Pre-training Data | Parameter Count | Status |
|---|---|---|---|---|
| 2D Graph | Mole-BERT | 2M molecules | - | Frozen |
| 3D Conformation | Uni-Mol | 209M conformations | - | Frozen |
| Image | CLIP (ViT) | LAION-400M (general domain) | - | Frozen |
| MS2 Spectrum | Transformer (SeqT) | Trained from scratch | - | Trainable |
| IR Spectrum | Transformer (SeqT) | Trained from scratch | - | Trainable |
Design Rationale: MS2 and IR encoders trained from scratch as Sequence Transformers treating spectral peaks as token sequences, since no suitable pre-trained models exist for chemical spectra.
Evaluation
Metrics:
- Accuracy (Acc) for recognition tasks
- BLEU-2/4 and METEOR for captioning tasks
- AUC-ROC for property prediction (classification)
- RMSE for property prediction (regression)
Benchmarks:
- ChemLLMBench: Adapted for multimodal inputs across molecule captioning, property prediction, and reaction understanding
- MoleculeNet: Standard molecular property prediction tasks (BACE, BBBP, HIV, ClinTox, SIDER, Tox21)
- USPTO: Reaction prediction and retrosynthesis tasks
- Custom Spectral Tasks: Novel evaluations requiring spectral interpretation
Hardware & Hyperparameters
Training Configuration:
- Total Batch Size: 256
- Epochs: 3
- Optimizer: AdamW
Modality-Specific Learning Rates (Peak):
| Modality | Learning Rate | Feature Dimension |
|---|---|---|
| Graph | 1e-5 | 300 |
| Conformation | 2e-4 | 512 |
| Image | 2e-3 | 1024 |
| MS2 / IR | 2e-4 | 768 |
Note: Different learning rates reflect the varying degrees of domain adaptation required - images (general CLIP) need more adaptation than graphs (chemical Mole-BERT).
Citation
@article{zhaoChemDFMXLargeMultimodal2024,
title = {{{ChemDFM-X}}: {{Towards Large Multimodal Model}} for {{Chemistry}}},
author = {Zhao, Zihan and Chen, Bo and Li, Jingpiao and Chen, Lu and Wen, Liyang and Wang, Pengyu and Zhu, Zichen and Zhang, Danyang and Wan, Ziping and Li, Yansi and Dai, Zhongyang and Chen, Xin and Yu, Kai},
year = {2024},
month = dec,
journal = {Science China Information Sciences},
volume = {67},
number = {12},
pages = {220109},
doi = {10.1007/s11432-024-4243-0},
archiveprefix = {arXiv},
eprint = {2409.13194},
primaryclass = {cs.LG}
}