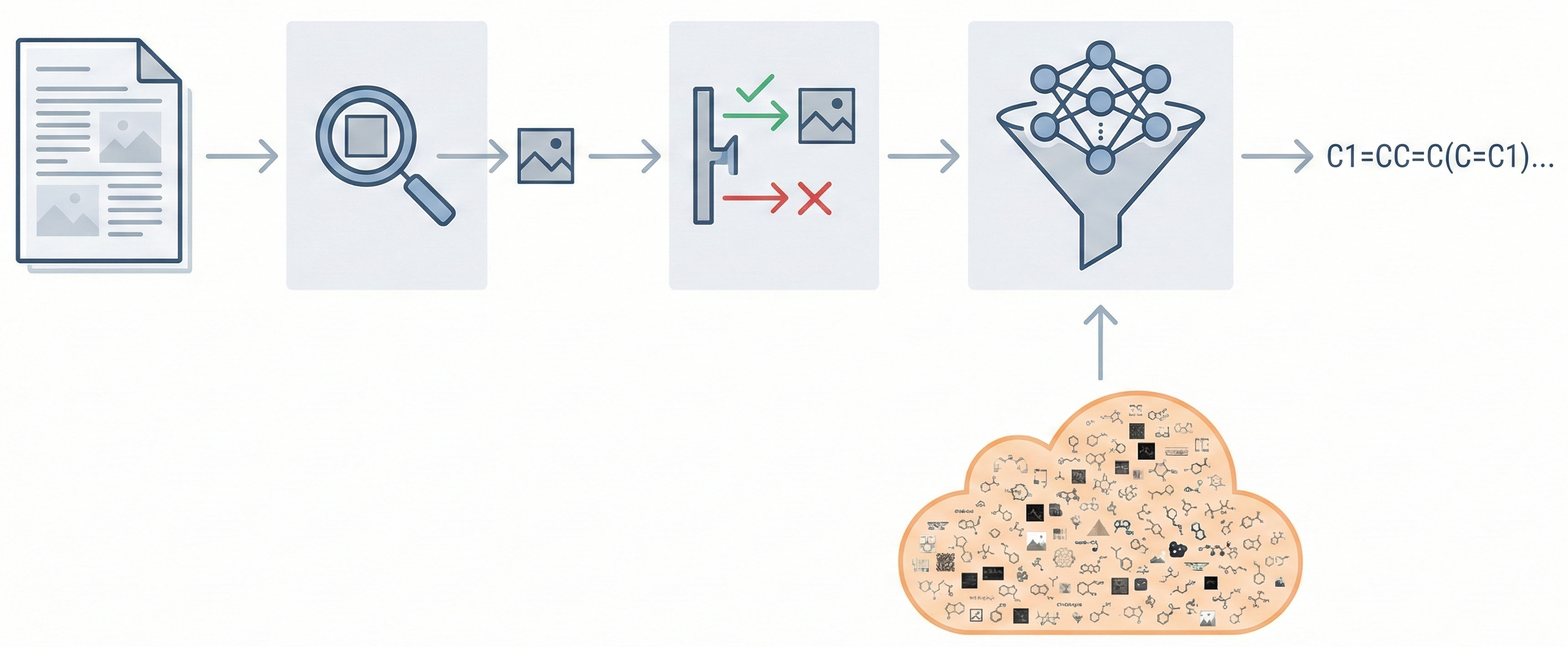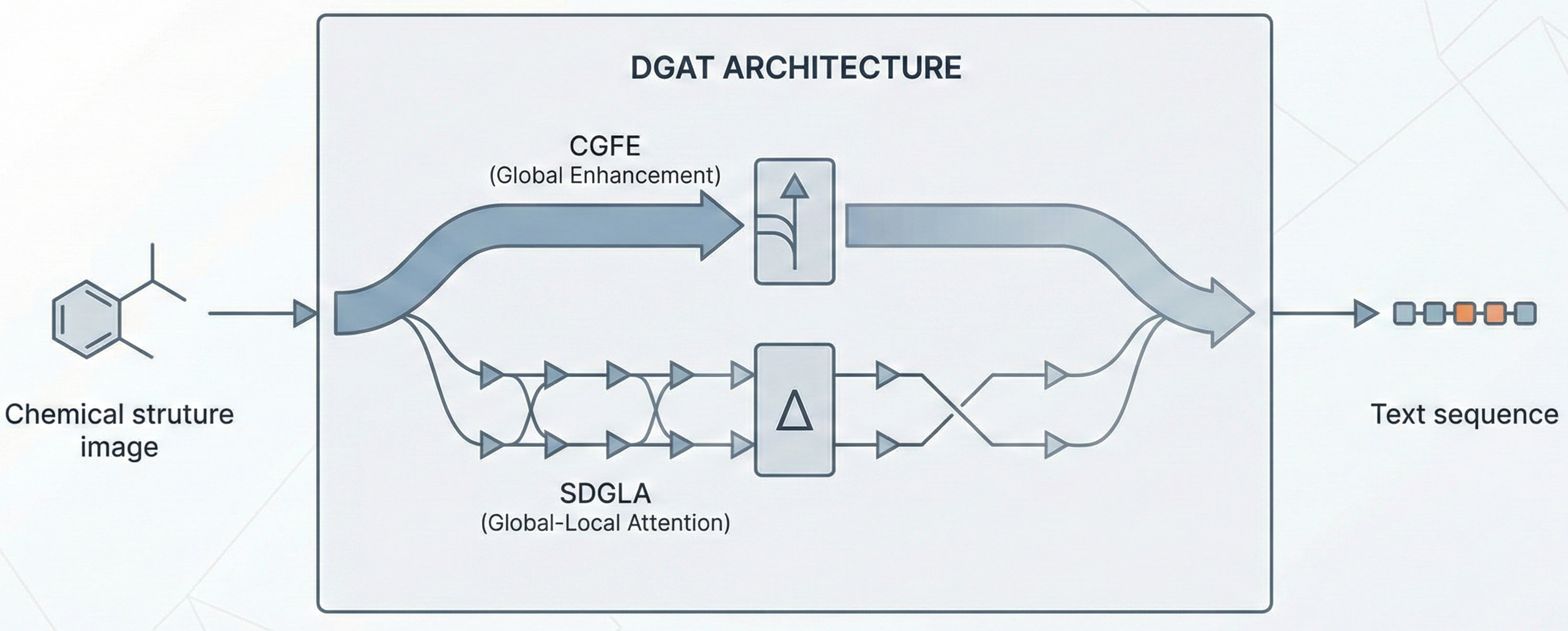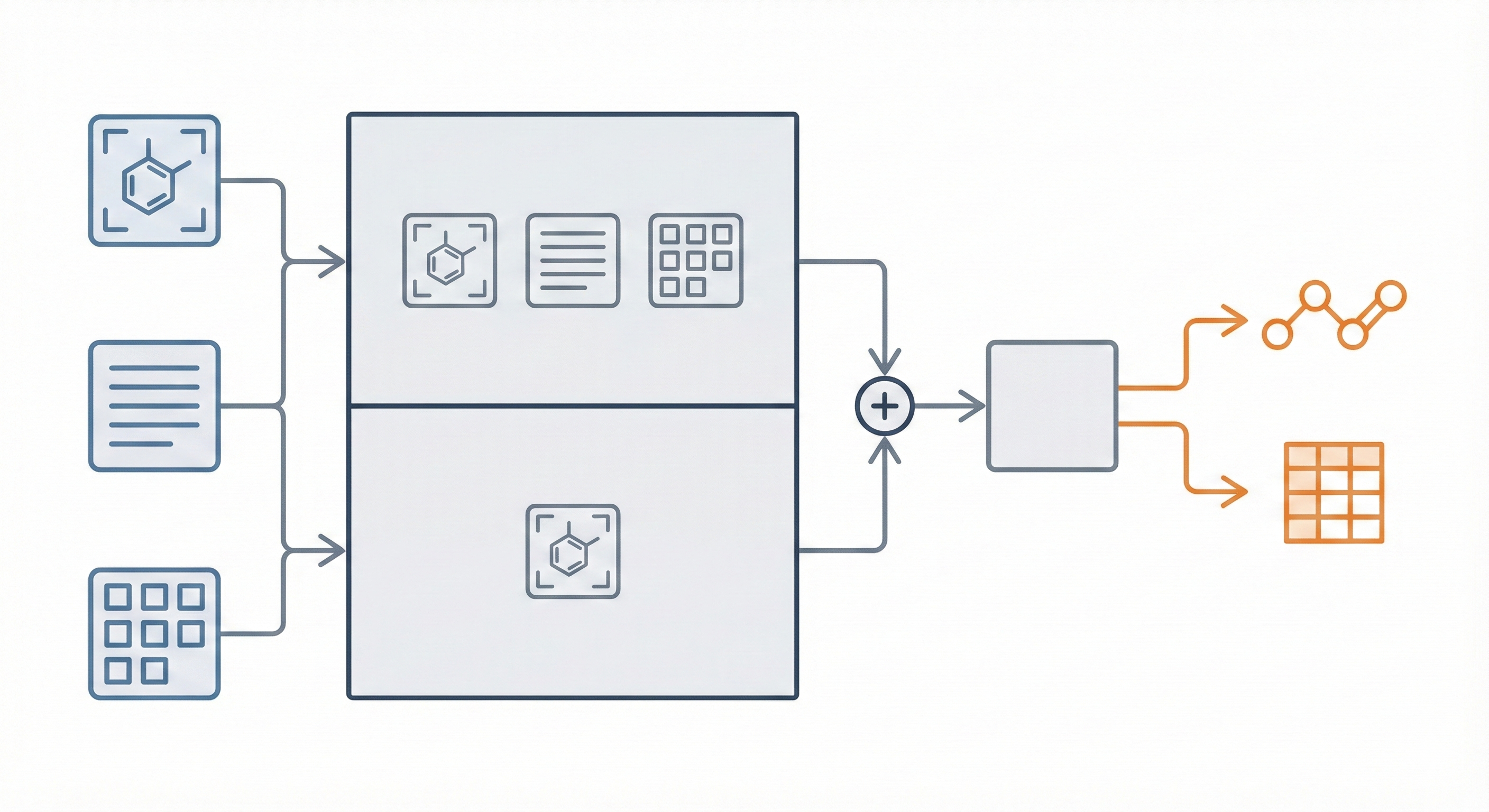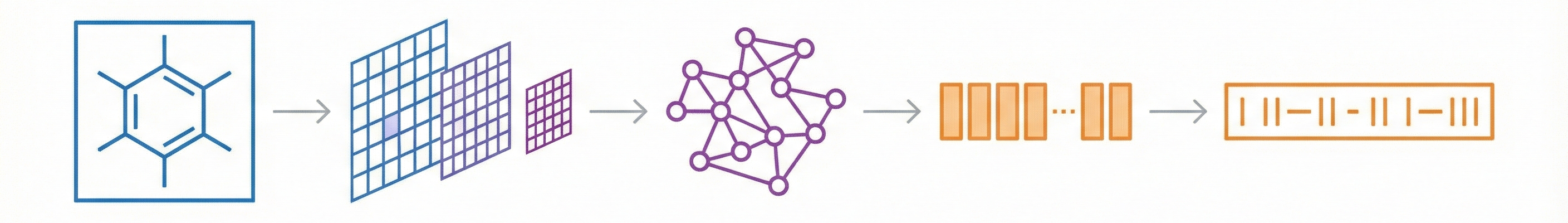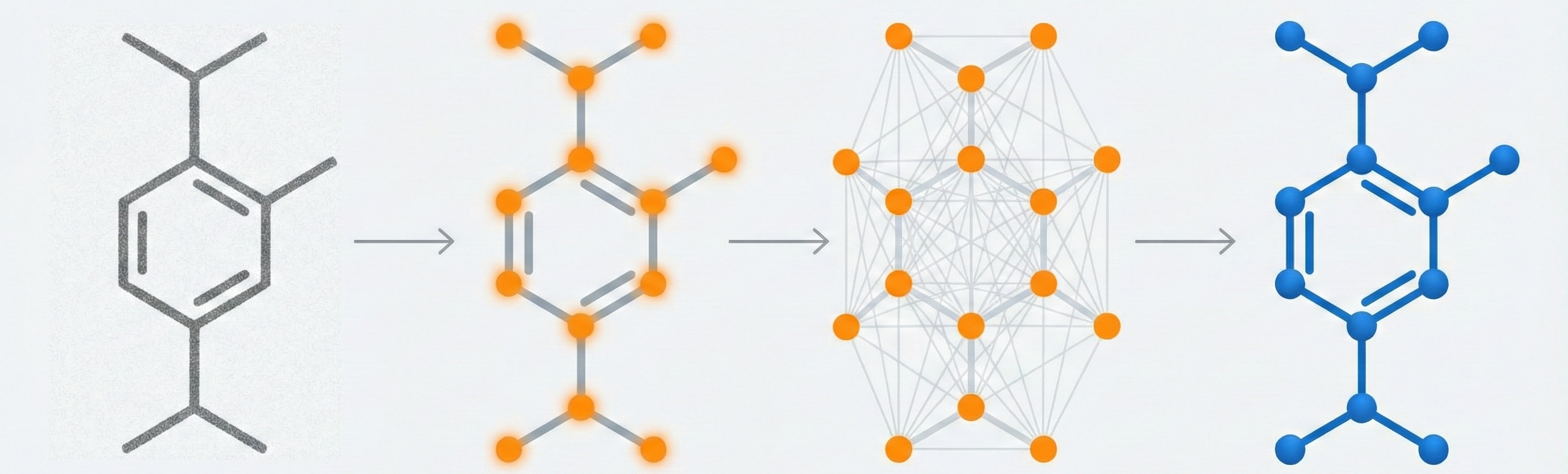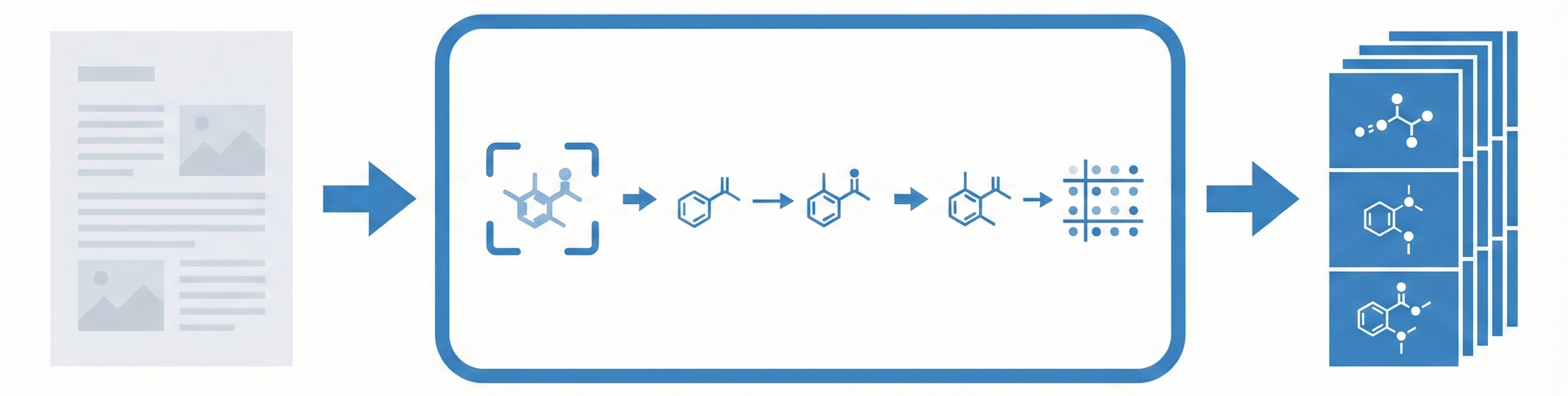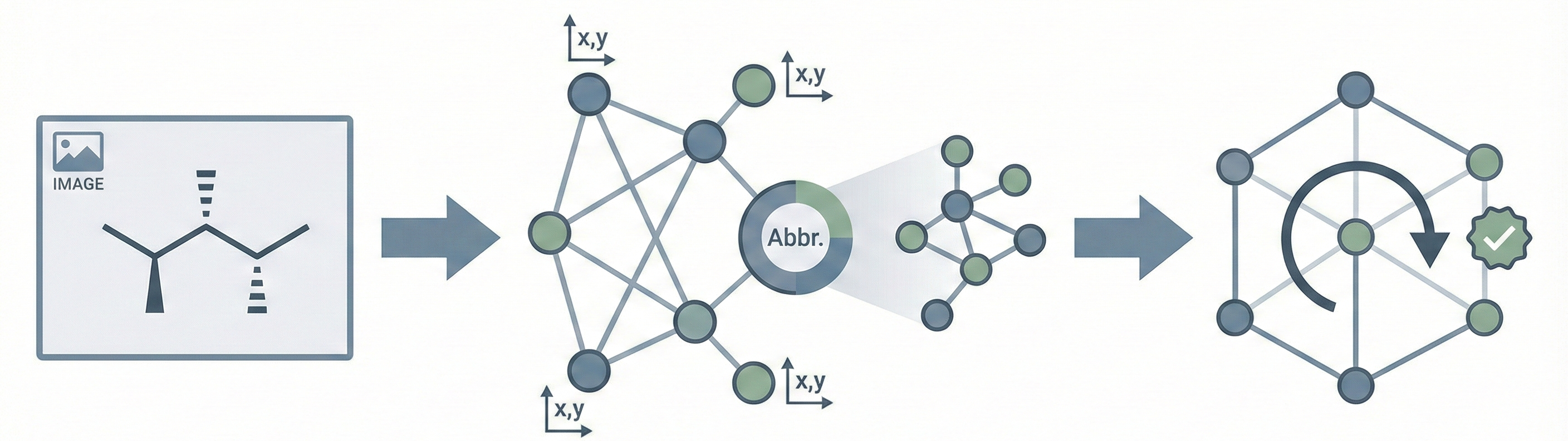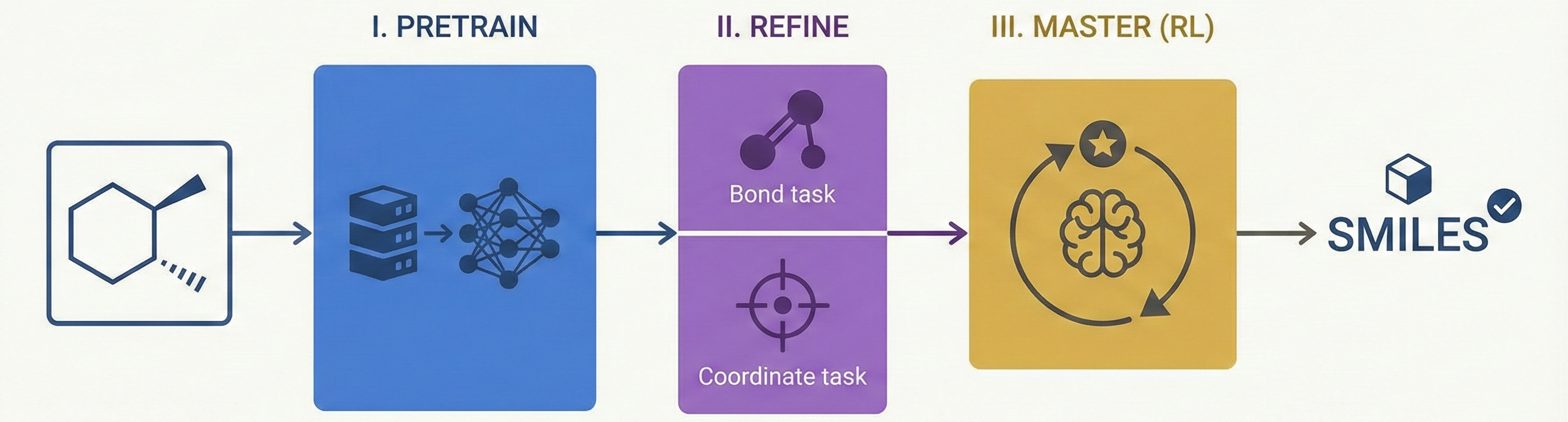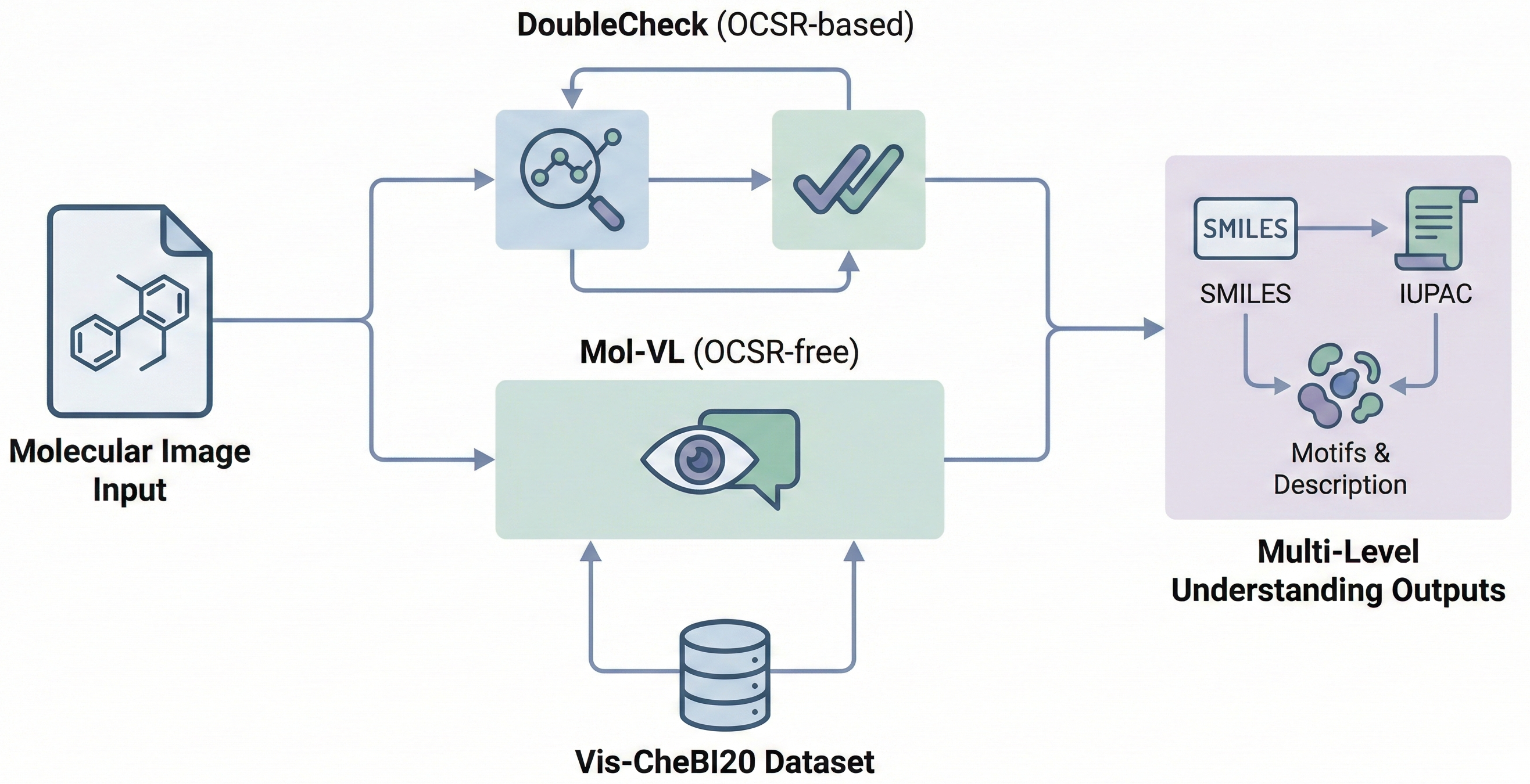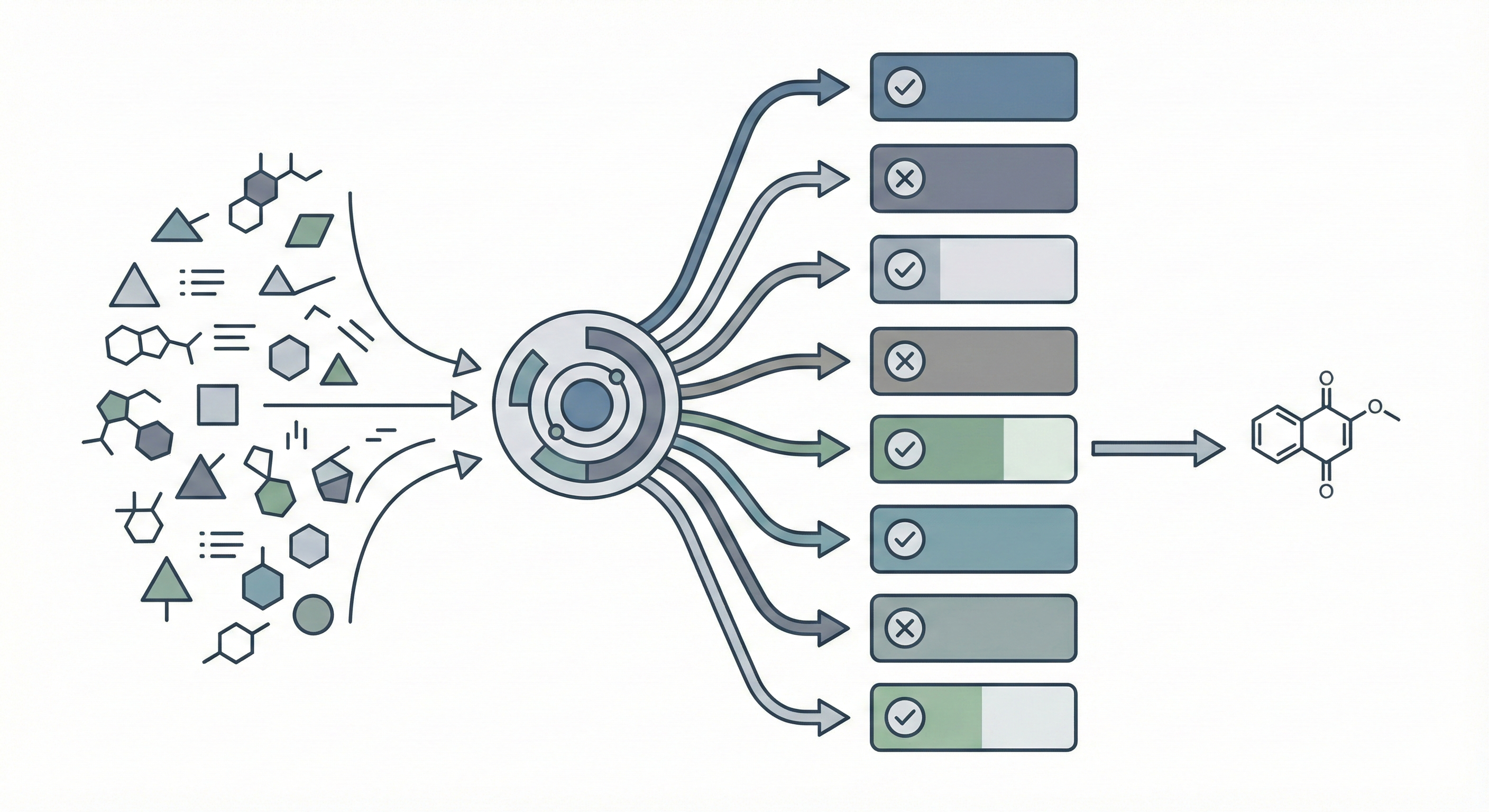
Comparing OCSR Tools (Krasnov et al. 2024)
Comprehensive evaluation of 8 optical chemical structure recognition tools using a newly curated dataset of 2,702 patent images. Proposes ChemIC, a ResNet-50 classifier to route images to specialized tools based on content type, demonstrating that no single tool excels at all tasks.