
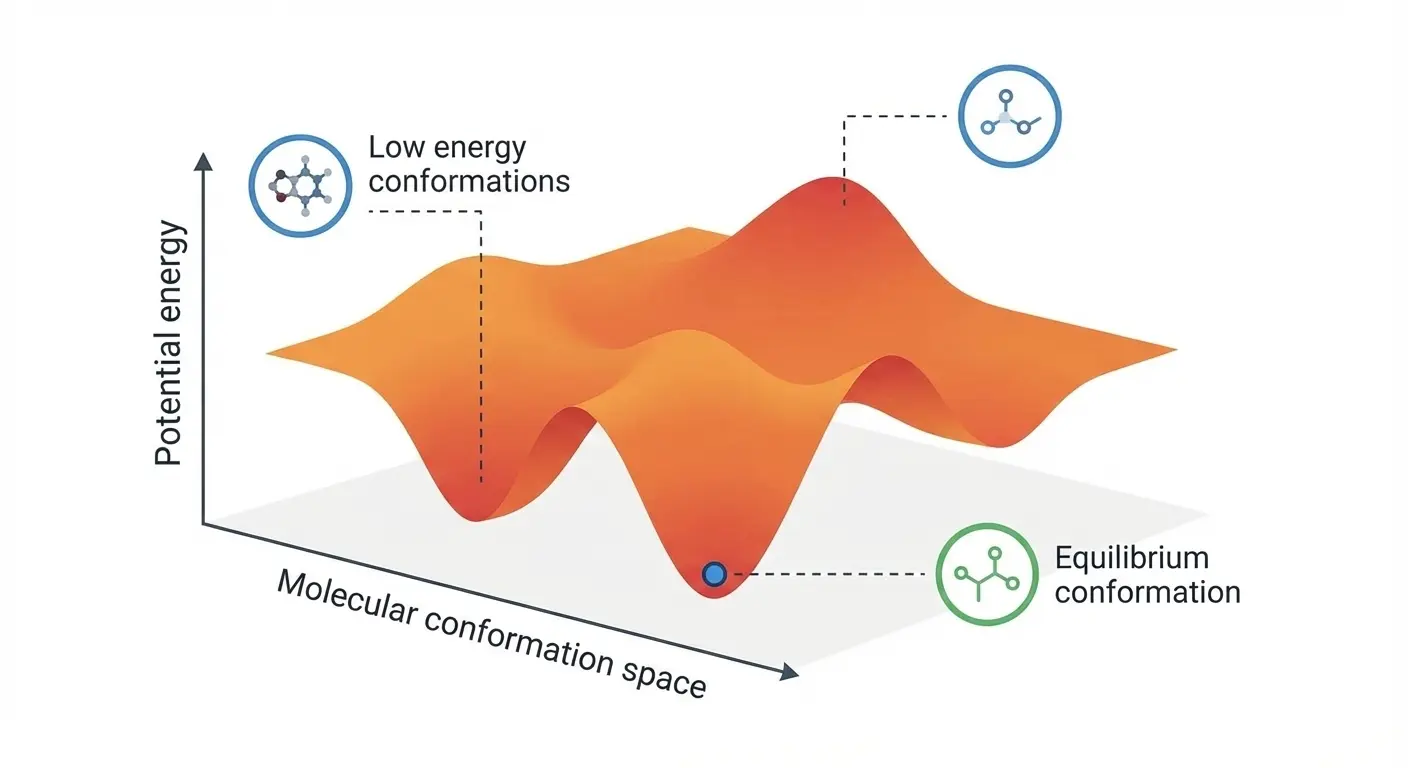
Müller-Brown Basin MB: Langevin Dynamics Simulation
Watch particle dynamics in the product minimum of the Müller-Brown potential. This simulation shows intermediate thermal motion behavior at -108.17 kJ/mol energy level.

Watch particle dynamics in the product minimum of the Müller-Brown potential. This simulation shows intermediate thermal motion behavior at -108.17 kJ/mol energy level.
ICLR 2025 paper introducing DenoiseVAE, which learns adaptive, atom-specific noise distributions through a VAE framework to improve denoising-based pre-training for molecular force field prediction, outperforming fixed Gaussian noise approaches on quantum chemistry benchmarks.
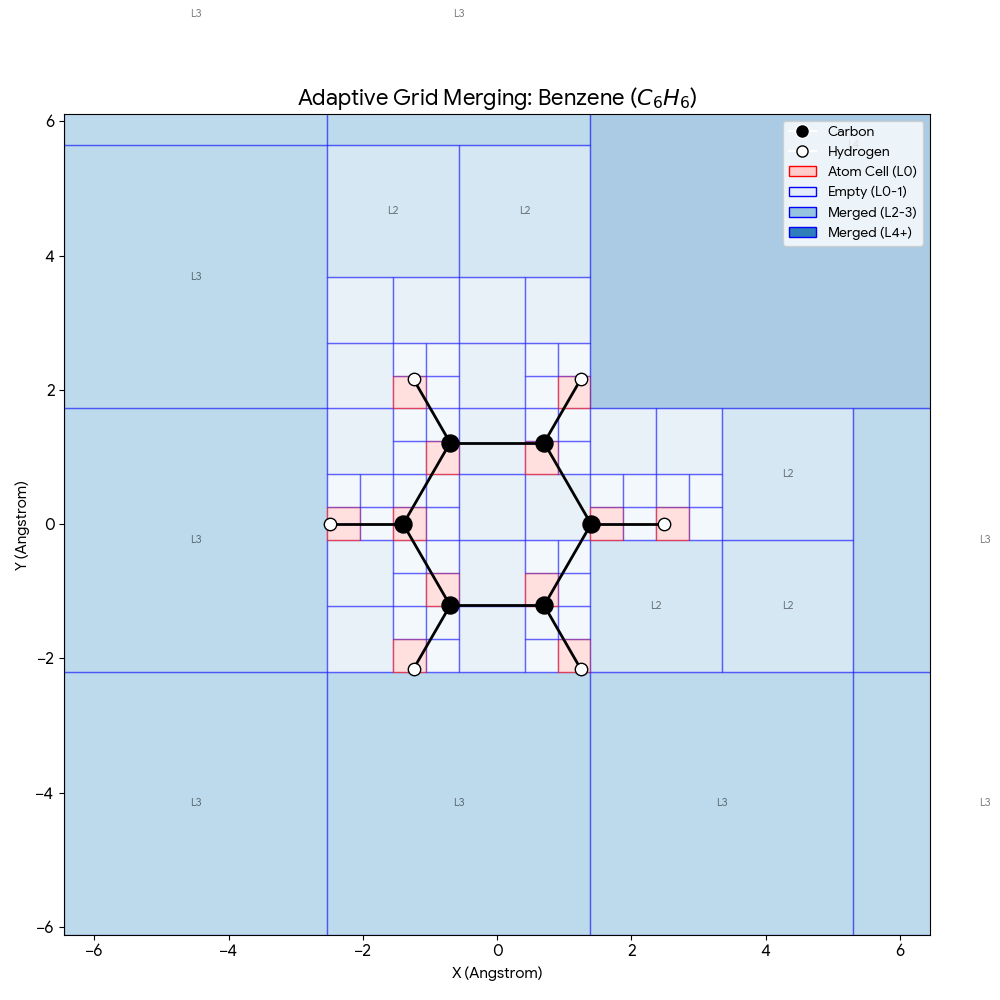
ICML 2025 paper introducing SpaceFormer, a Transformer architecture that challenges the atom-centric paradigm by modeling the continuous 3D space surrounding molecules using adaptive multi-resolution grids, achieving state-of-the-art performance on quantum property prediction benchmarks.
ICML 2025 analysis rigorously quantifying when non-conservative force models (which predict forces directly) fail in molecular dynamics, demonstrating simulation instabilities and proposing hybrid architectures that capture speed benefits without sacrificing physical correctness.
ICML 2025 methodological paper introducing SPHNet, which uses adaptive network sparsification to overcome the computational bottleneck of tensor products in SE(3)-equivariant networks, achieving 2-5x speedup while maintaining accuracy on DFT Hamiltonian prediction tasks.

ICML 2025 paper proposing energy conservation metrics as critical diagnostics for machine learning interatomic potentials and introducing eSEN, a novel architecture designed to bridge the gap between test-set accuracy and real simulation performance on materials property prediction.
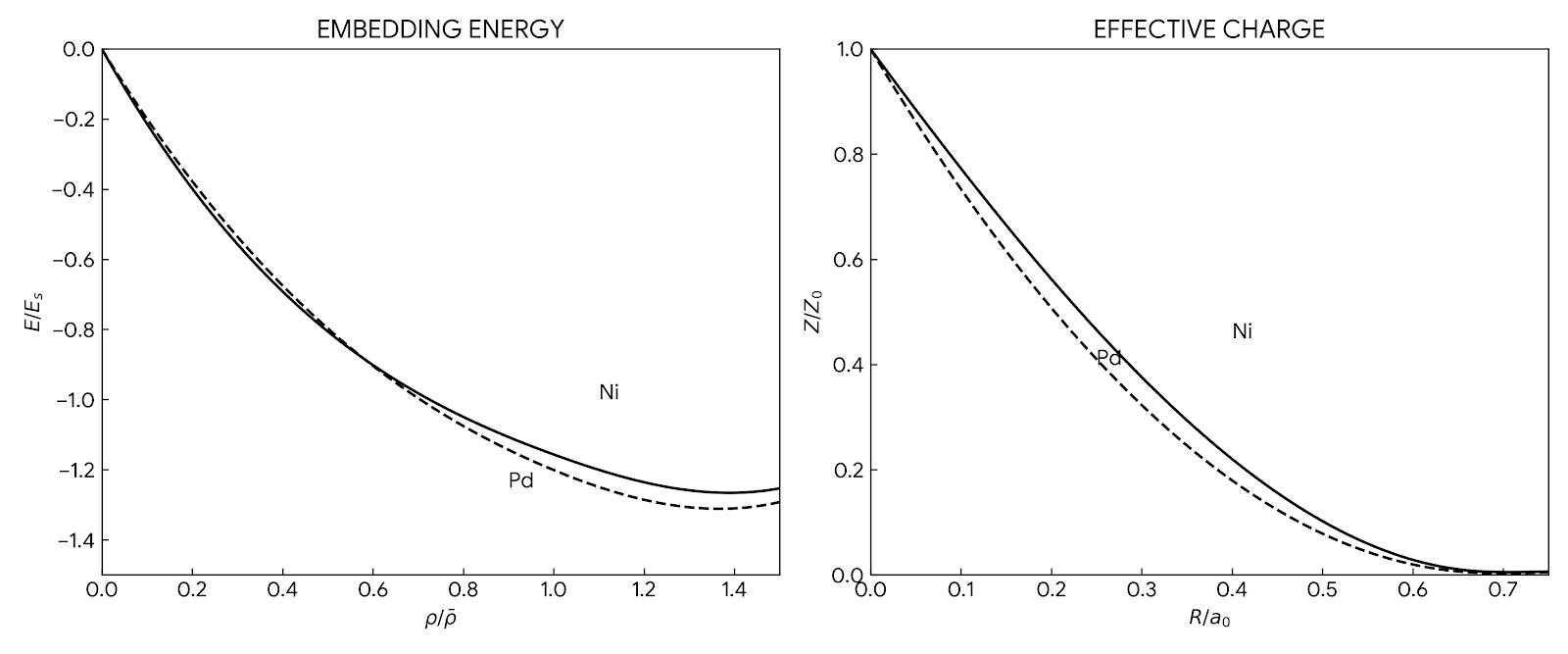
The foundational 1984 paper introducing EAM, a semi-empirical many-body interatomic potential that incorporates density functional theory concepts to accurately simulate metallic systems while maintaining computational efficiency comparable to pair potentials.
Torrie and Valleau’s 1977 methodological breakthrough introducing importance sampling with non-physical distributions to overcome the sampling gap problem in Monte Carlo free-energy calculations, particularly for rare events and phase transitions.
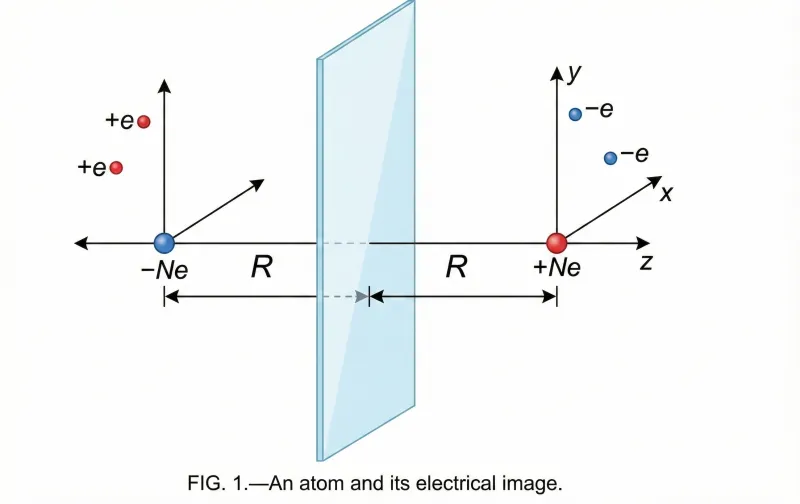
Lennard-Jones’s landmark 1932 theoretical paper that applied quantum mechanical potential energy surfaces to gas-solid interactions, providing the first unified framework explaining both physisorption and chemisorption as different regions of the same energy landscape.
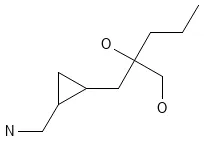
GDB-13 contains nearly 1 billion systematically generated small organic molecules with up to 13 atoms, achieving billion-scale chemical space exploration while maintaining drug-like properties.
GDB-17 contains 166 billion systematically generated small organic molecules with up to 17 atoms, representing the most comprehensive exploration of drug-relevant chemical space achieved through computational enumeration.
Get a practical overview of the GEOM dataset and learn how it’s advancing 3D molecular machine learning by bridging static graphs and dynamic reality.