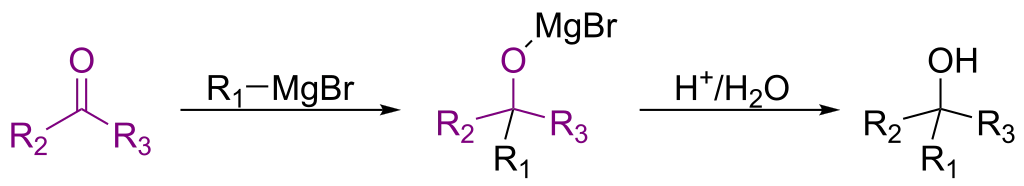
RInChI: Reaction International Chemical Identifier
A 2018 infrastructure paper introducing RInChI (Reaction InChI), the first standardized format for uniquely identifying chemical reactions through algorithmic hashing and layering, enabling reaction database searching and duplicate detection analogous to how InChI works for individual molecules.