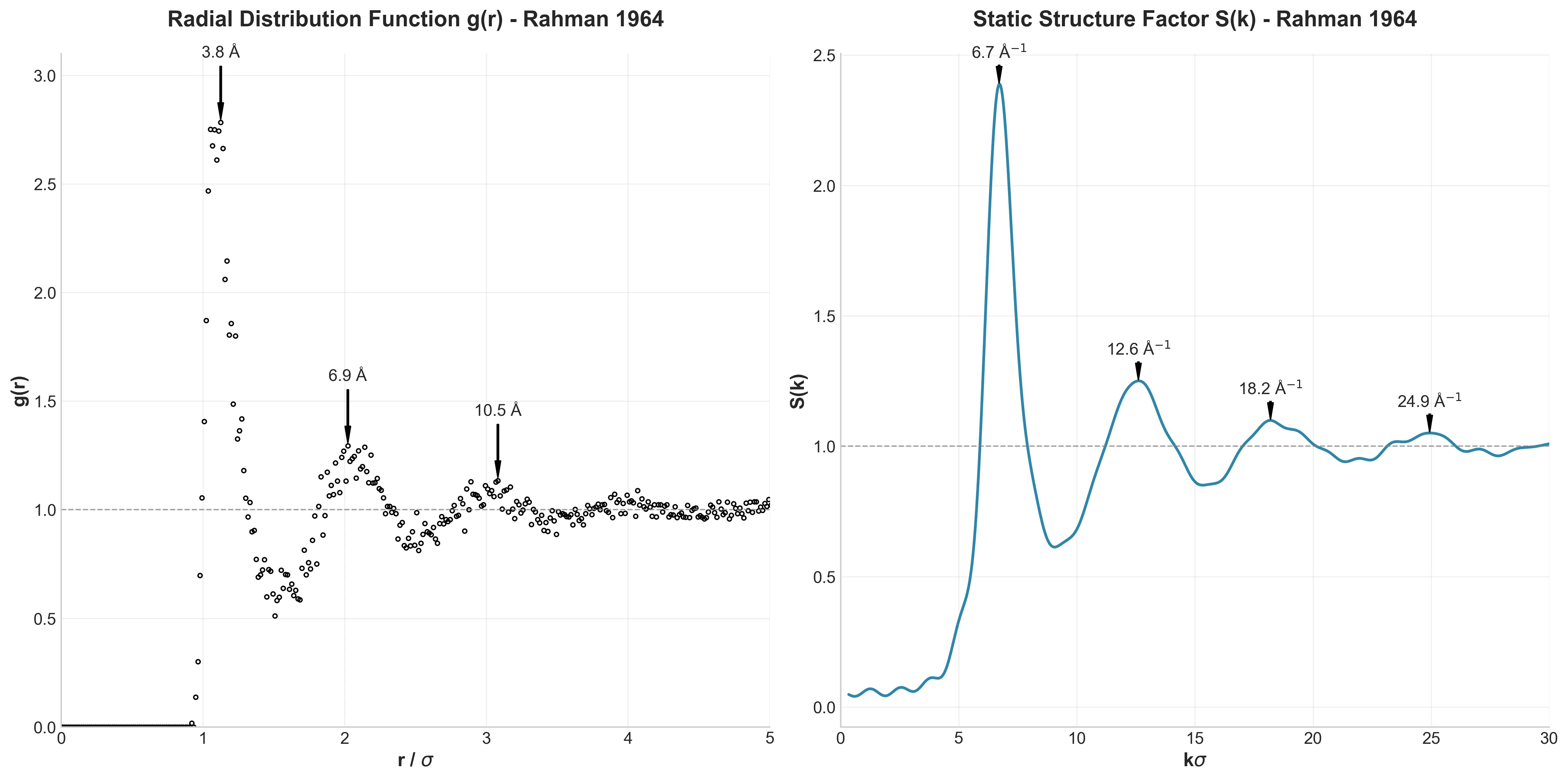
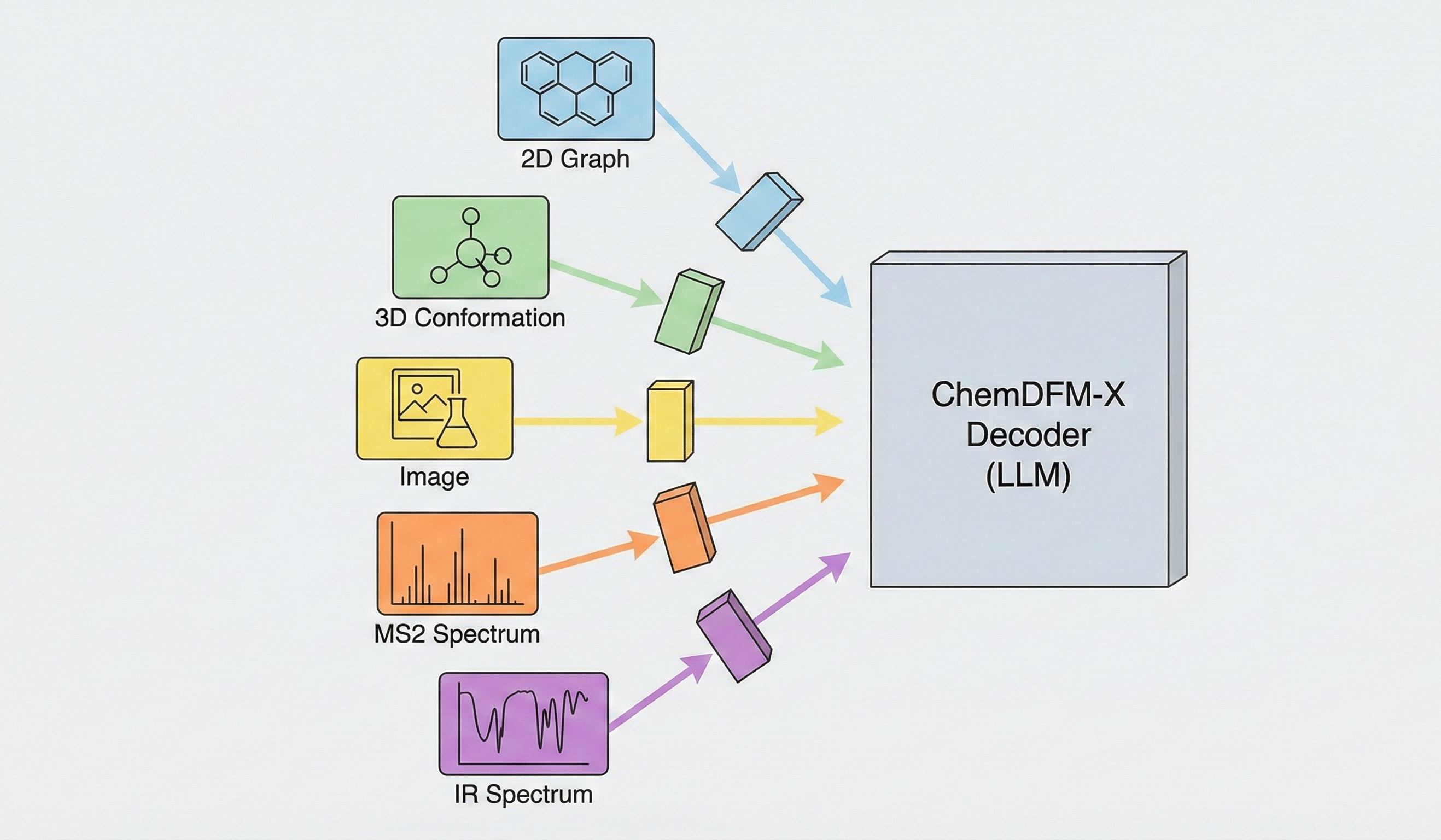
Müller-Brown Potential: A PyTorch ML Testbed
A high-performance, GPU-accelerated PyTorch testbed for ML-MD algorithms featuring JIT-compiled analytical Jacobian force kernels achieving 3-10x speedup over autograd, robust Langevin dynamics with Velocity-Verlet integration, and modular architecture designed as ground-truth validation for novel machine learning approaches in molecular dynamics.