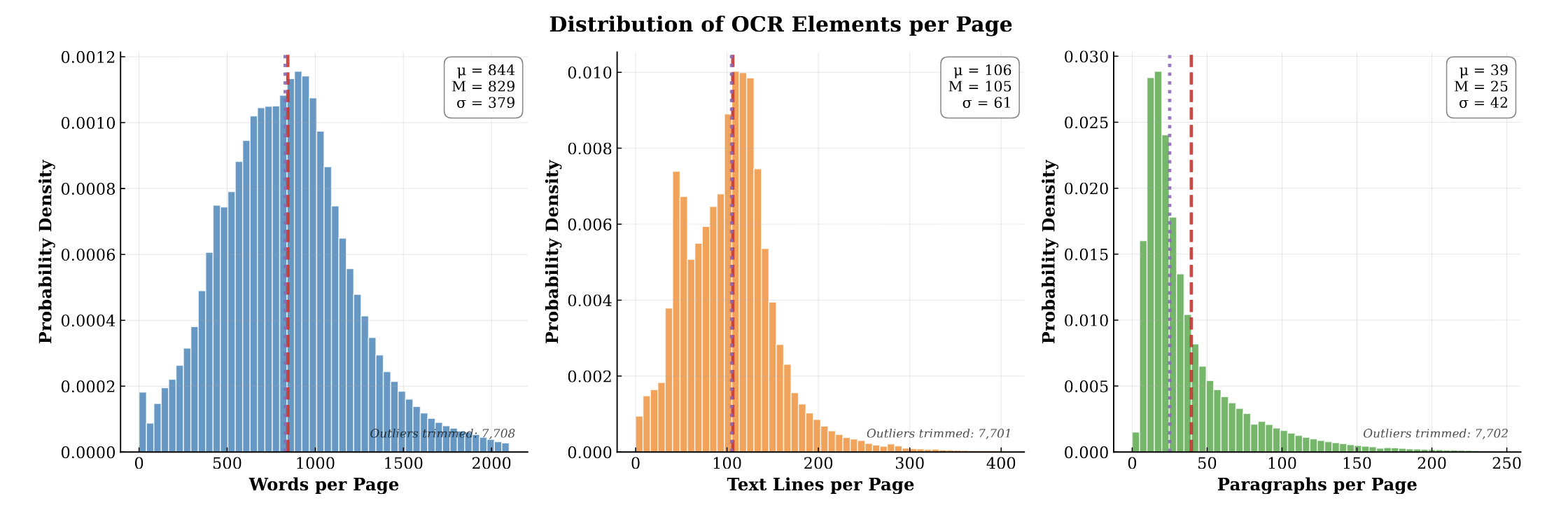
PubMed-OCR: PMC Open Access OCR Annotations
A large-scale dataset of 209K+ articles with OCR and layout bounding boxes, enabling layout-aware modeling and document …

A large-scale dataset of 209K+ articles with OCR and layout bounding boxes, enabling layout-aware modeling and document …
MOSES provides a standardized benchmarking platform for molecular generative models, featuring datasets, metrics, and …
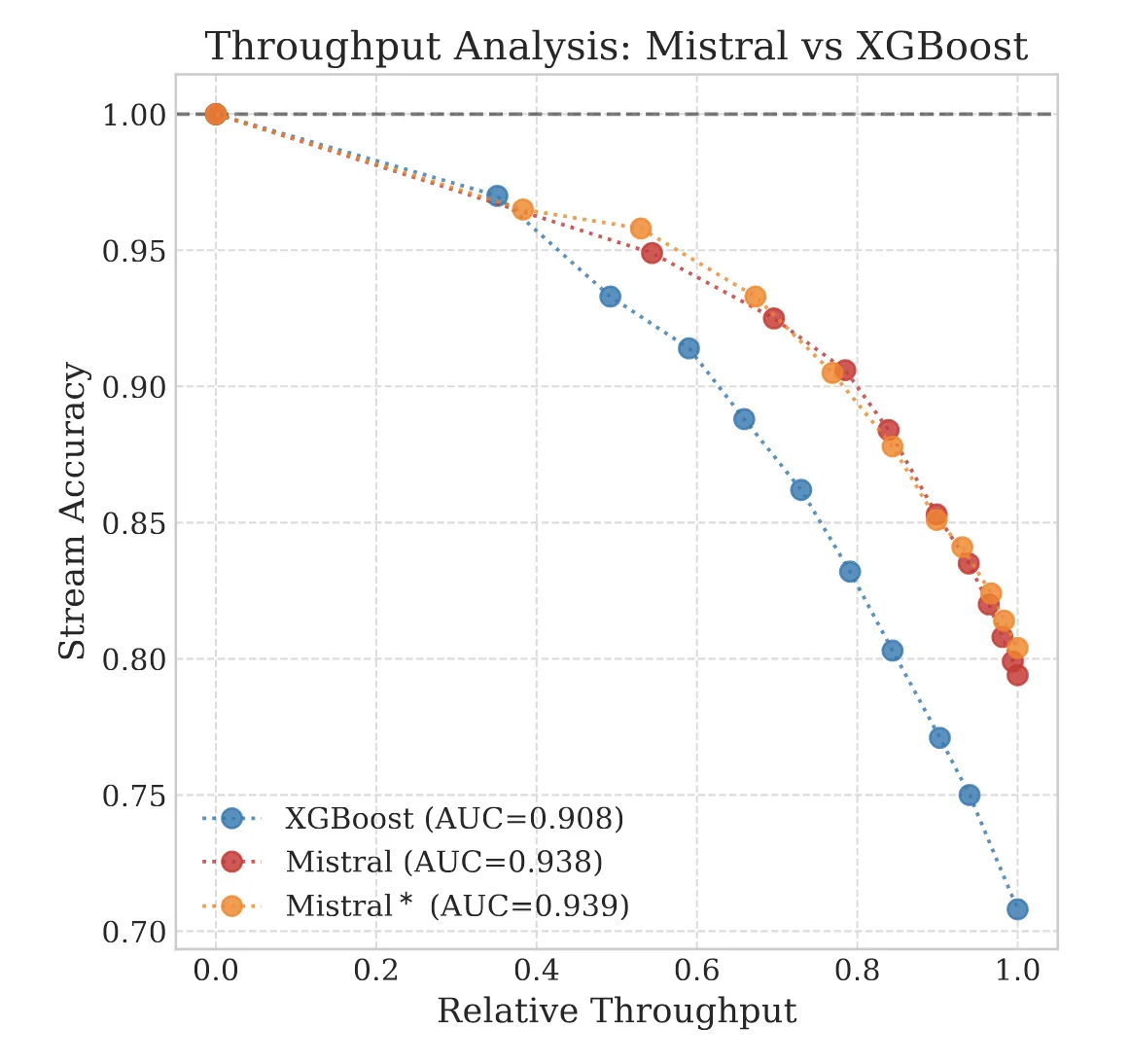
Why high-accuracy LLMs fail in production: exploring the calibration crisis and the challenge of reliable …
From brittle rules to reasoning engines: why transformers are the only way to solve the 'Hello World' of document …

Optimizing transformer pretraining for molecules using MLM vs MTR objectives, scaling to 77M compounds from PubChem for …
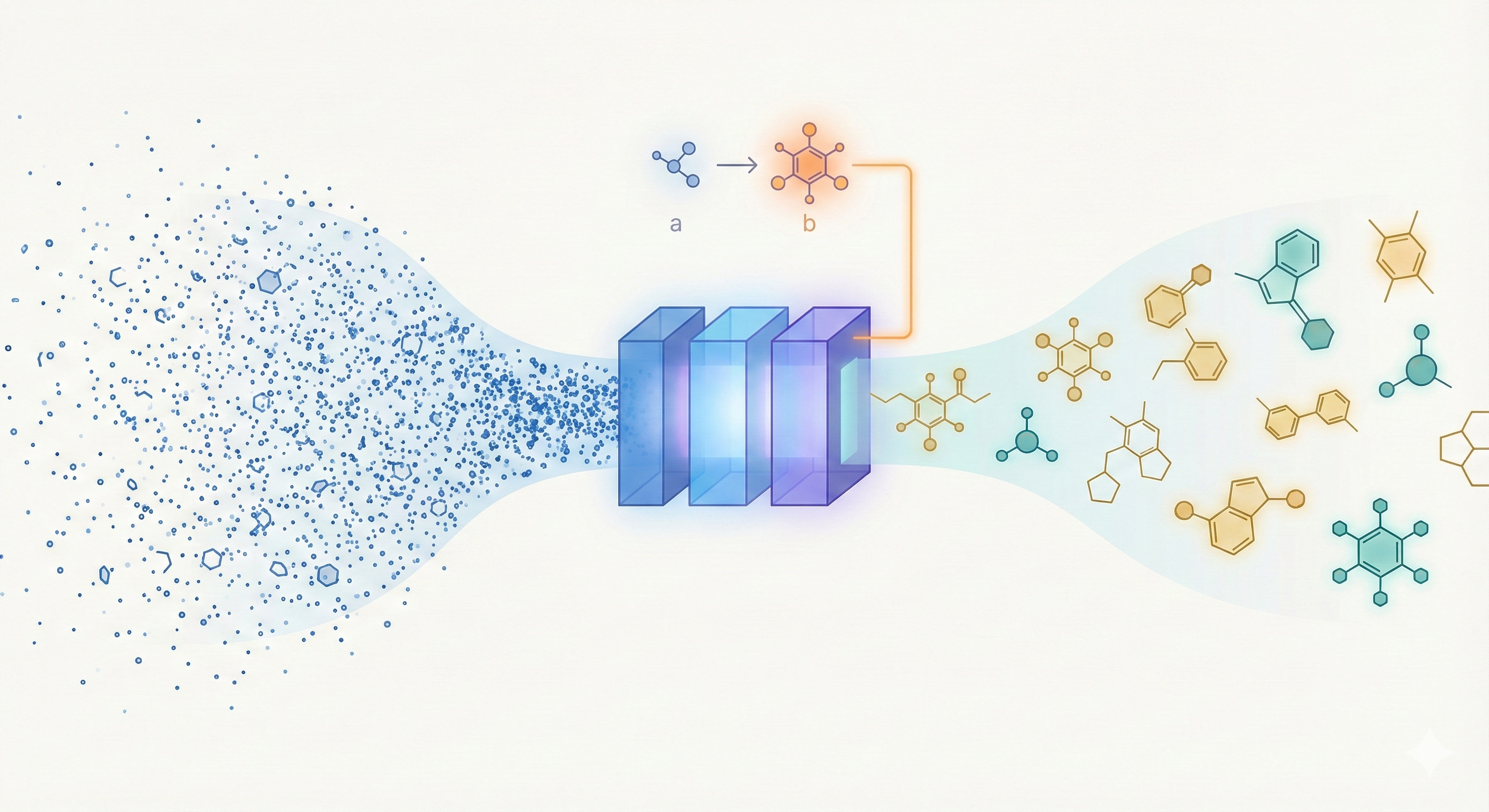
A 46.8M parameter transformer for molecular generation trained on 1.1B SMILES, introducing pair-tuning for efficient …

A systematic evaluation of RoBERTa transformers pretrained on 77M PubChem SMILES for molecular property prediction …
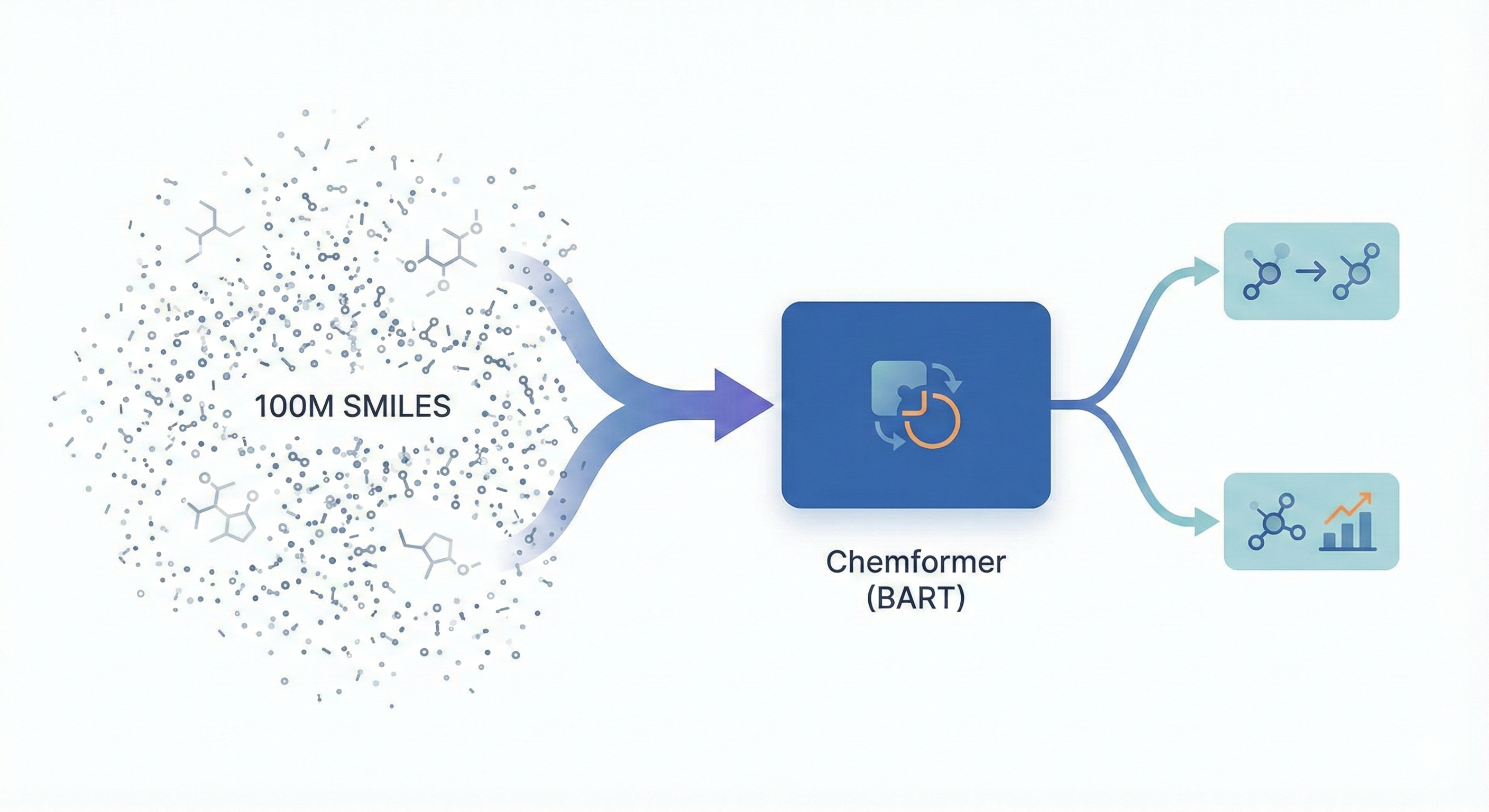
BART-based Transformer pre-trained on 100M molecules using self-supervision to accelerate convergence on chemical …
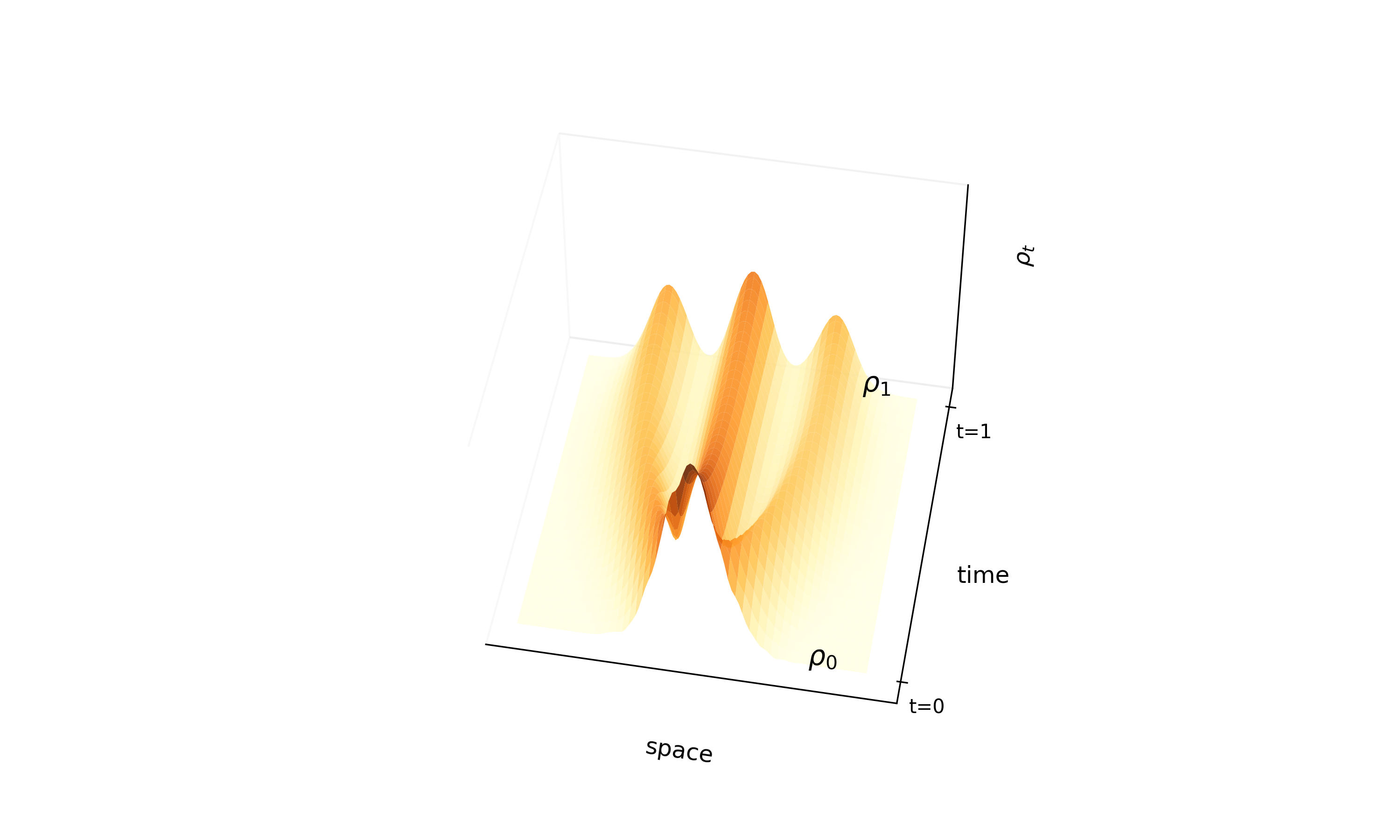
A continuous-time normalizing flow using stochastic interpolants and quadratic loss to bypass costly ODE …
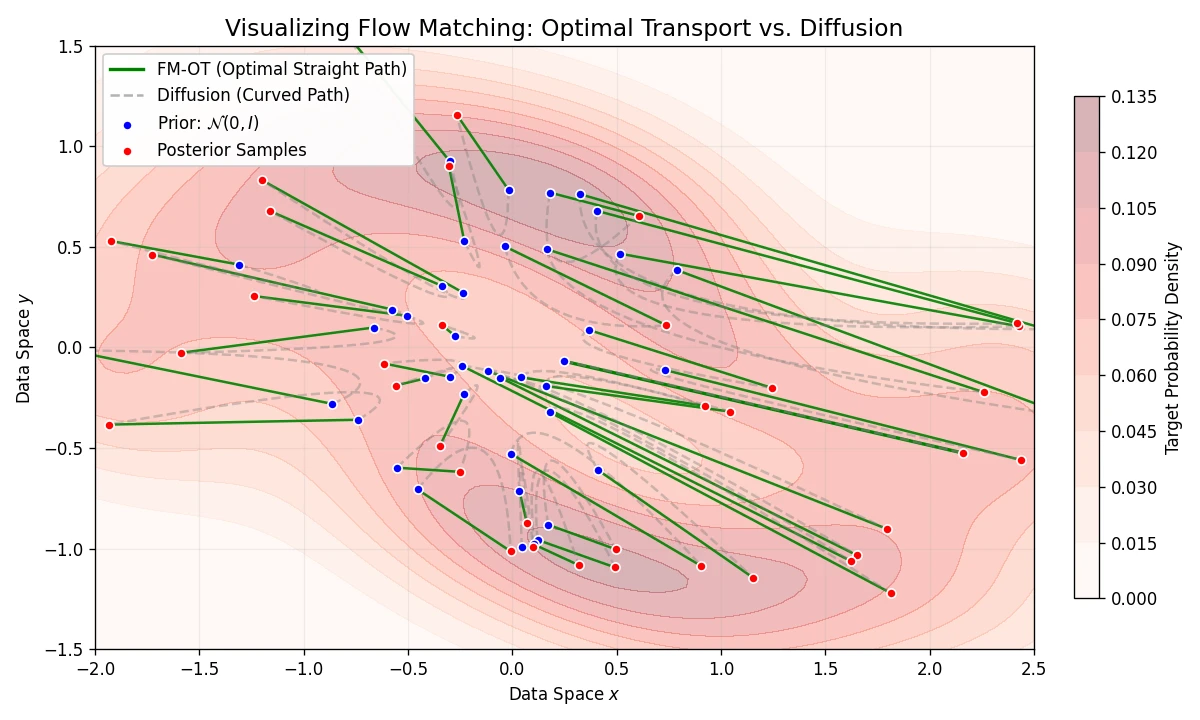
A simulation-free framework for training Continuous Normalizing Flows using Conditional Flow Matching and Optimal …
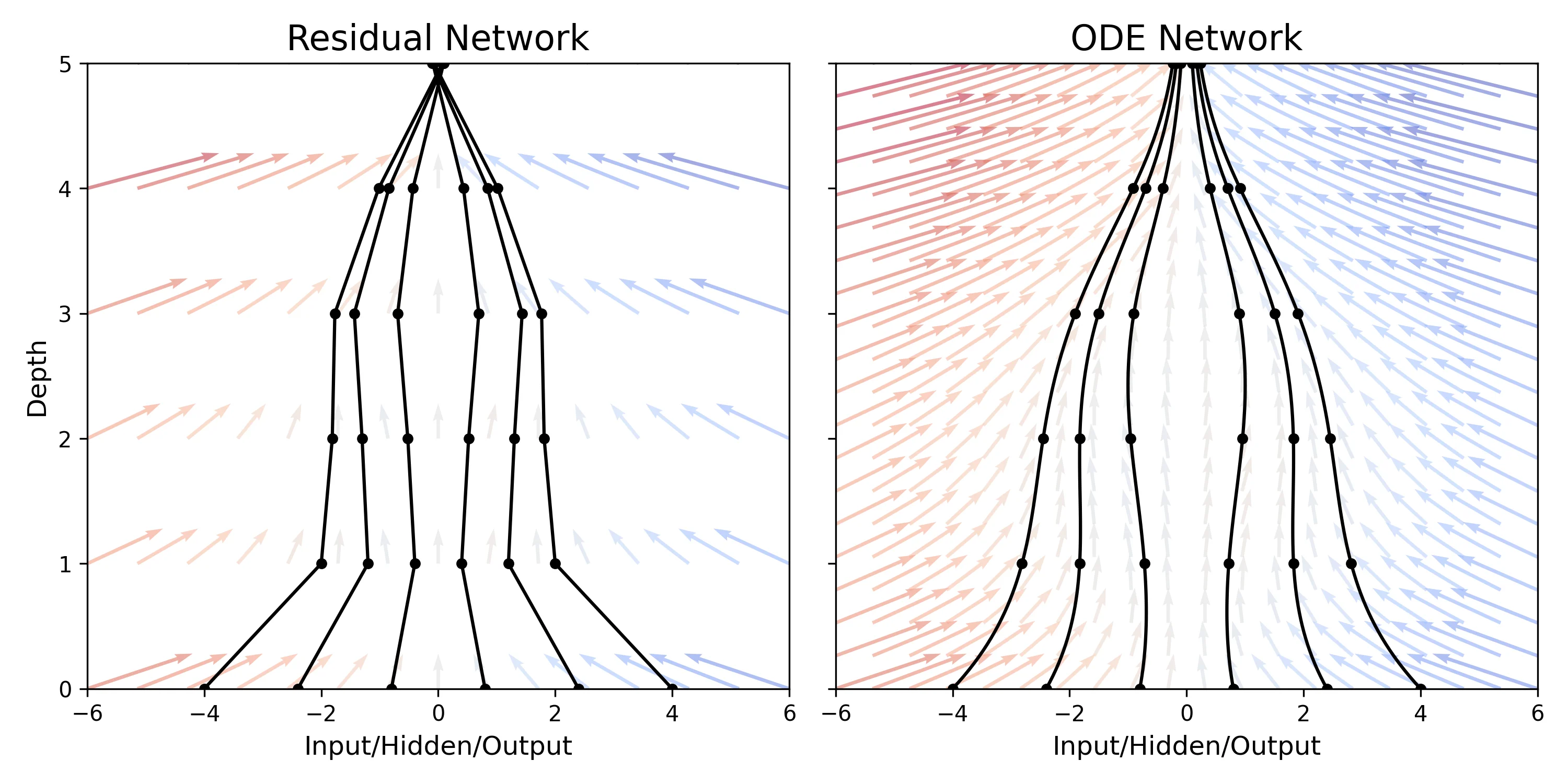
Introduces ODE-Nets, a continuous-depth neural network model parameterized by ODEs, enabling constant memory …
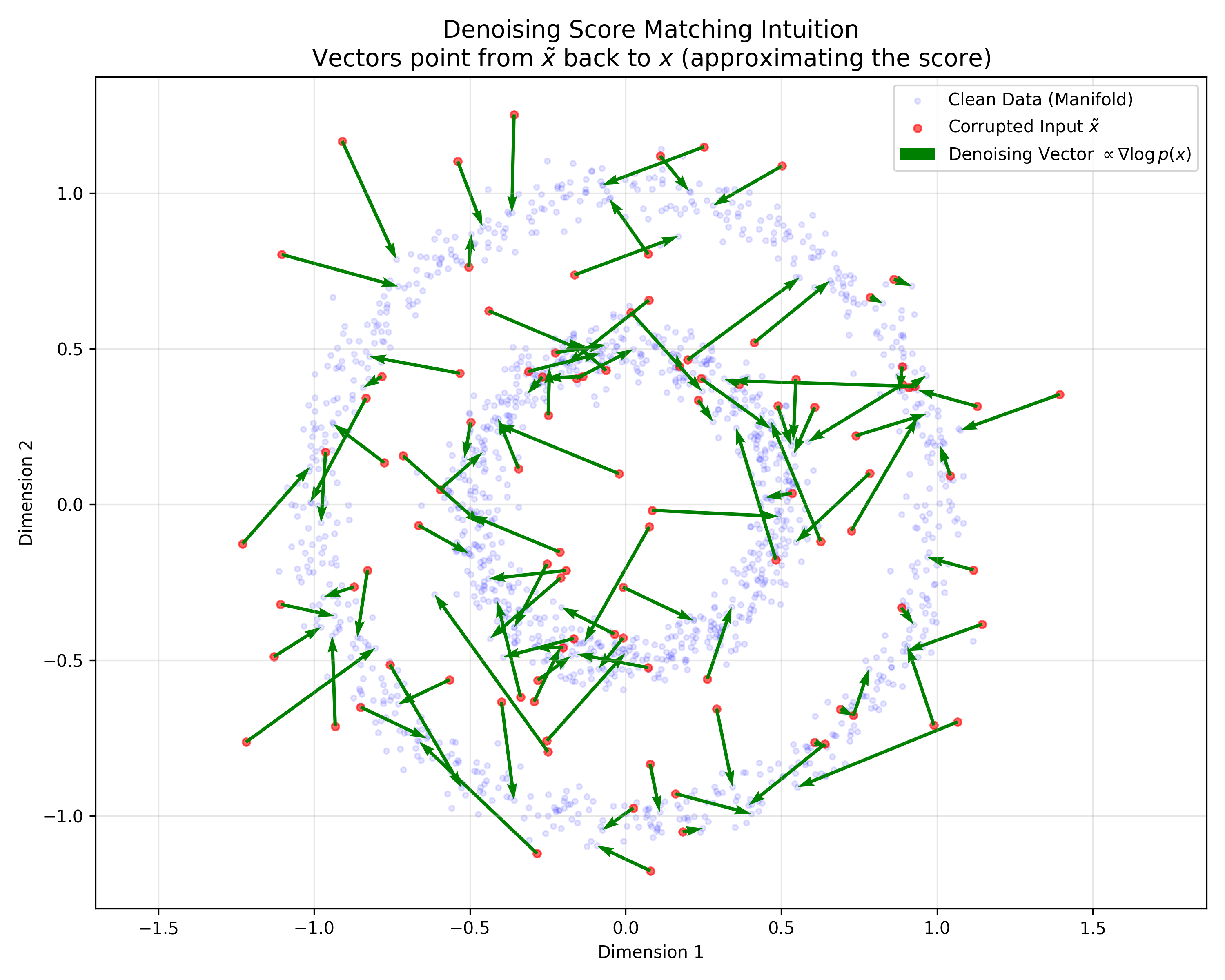
Theoretical paper proving the equivalence between training Denoising Autoencoders and performing Score Matching on a …